This tree corresponds to the best maximum likelihood tree derived using a 7,160 site alignment and the GTRCAT model in RAxML99. The tree contains 16,821 OTUs generated from PacBio sequencing of 21 environmental samples (no reference sequences were included). ring no. 1 around the tree indicates the taxonomy of environmental sequences, labeling all major eukaryotic lineages considered in this study. ring no. Figure 2 represents the percent similarity to the references in the PR2 database as calculated using BLAST and was set at a minimum of 70%, with the two black lines in the middle indicating 85% and 100% similarity levels. ring no. 3 shows the habitat origin of each OTU. b, Hierarchical clustering of the four habitats based on a phylogenetic distance matrix constructed using the unweighted UniFrac method (n=7, n=5, n=4, and n=5 samples for soil, freshwater, marine euphotes, and marine aphotes, respectively ) . All communities were found to be significantly different from each other using Monte Carlo simulations (Bonferroni-adjusted P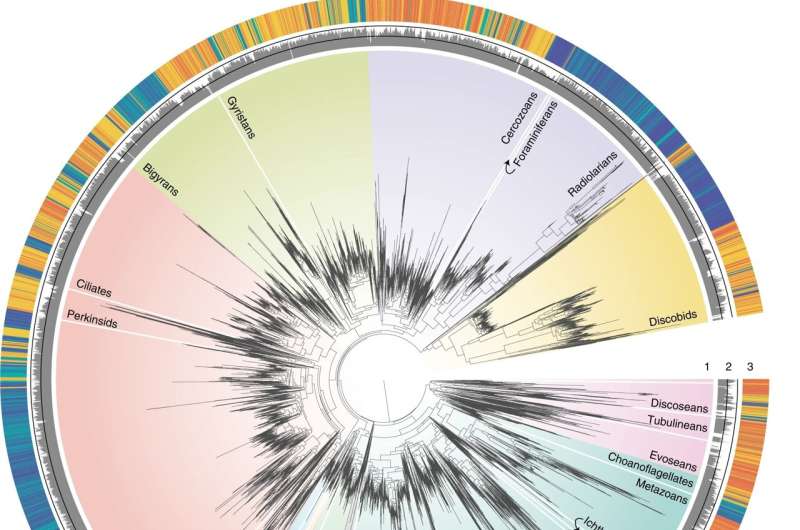
A recently published study in natural ecology and evolution has revealed some of the key processes in marine microbial evolution. According to the study, led by Uppsala University (Sweden) and involving the Institut de Ciències del Mar (ICM-CSIC) from Barcelona, it is the large number of habitat transitions – from sea to land and vice versa – that have occurred over the past million years which explains the great diversity today.
According to the authors, “crossing the salinity limit is not easy for organisms, and when it does, the resulting transitions are important evolutionary events that can trigger explosions of diversity.” which includes animals, plants and a variety of eukaryotic microorganisms.
Small but very versatile
In particular, the work now published has shown that microbial eukaryotes have made hundreds of large leaps from sea to land and also to freshwater habitats and vice versa during their evolution. This in turn has made it possible to deduce where the ancestors of each of the eukaryotic microbial groups were found.
“Thanks to the fact that we have good phylogenetic trees and samples from different environments, we were able to analyze the habitat transitions in different groups of eukaryotes that took place hundreds of times over millions of years of eukaryotic evolution, which is more than we think,” explains Ramon Massana, ICM-CSIC researcher and one of the authors of the study.
For its elaboration, the scientific team used the latest technologies to sequence the DNA of microbes living in samples collected from boreal lakes, forest floors, the Indian Ocean and the Mariana Trench, among others. In particular, the ICM-CSIC provided marine samples collected during the Malaspina expedition at different oceans and water column depths.
Thanks to this, it was possible to construct large evolutionary trees of the organisms found in these environments and even observe a number of patterns in the evolution of habitat preferences.
“We found that organisms in the eukaryotic tree of life are generally grouped according to whether they live in oceans or non-marine habitats,” explains Mahwash Jamy, a researcher at Uppsala University and lead author of this study. In this context, Jamy adds that “this finding confirms that adapting to a different salinity – or crossing the salt barrier – is difficult even for microbes.”
Nevertheless, the study proves that microbial eukaryotes have successfully established themselves in new habitats hundreds of times during their evolution. Therefore, the authors propose that it was precisely these elusive transitions that would have allowed colonizing organisms to occupy unoccupied ecological niches, leading to the great diversity of eukaryotes today.
Further notes on the first eukaryotes
On the other hand, evolutionary trees constructed from DNA sequences have also allowed researchers to zoom into the deep past and deduce what the habitats of the ancestors of each microbial group might have been.
“It is likely that two of the largest groups of eukaryotes, the SARS and the Obozoa, each larger than say animals or plants, arose in completely different habitats,” says Fabien Burki, also a researcher at Uppsala University and another Lead author of the study.
According to Burki, the SAR lineage – which includes groups such as diatoms, ciliates, dinoflagellates, radiolarians, etc. – would have first emerged in the Precambrian oceans, while the ancestor of the Obazoa group – which diversified into fungi, animals, choanoflagellates, and amoebas – could have lived in non-marine habitats.”
This shows once again that crossing the salinity limit has played an important role in shaping eukaryotic evolution. For this reason, future research experts will turn to genomics to understand the genetic mechanisms underlying these key evolutionary events.
Chromatin is said to have originated in ancient microbes one to two billion years ago
Mahwash Jamy et al, Global Patterns and Rates of Habitat Transition in the Eukaryotic Tree of Life, Natural Ecology & Evolution (2022). DOI: 10.1038/s41559-022-01838-4
Provided by the Institut de Ciències del Mar
Citation: Researchers unveil key processes in marine microbial evolution (2022, August 5), retrieved August 5, 2022 from https://phys.org/news/2022-08-unveil-key-marine-microbial-evolution.html
This document is protected by copyright. Except for fair trade for the purpose of private study or research, no part may be reproduced without written permission. The content is for informational purposes only.
#Researchers #unveil #key #processes #marine #microbial #evolution

Leave a Comment