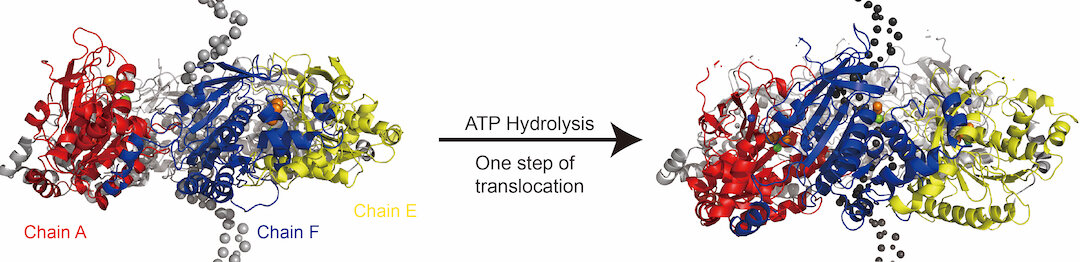
A helicase protein model created at Rice University shows a before-and-after picture of how the six-sided ring moves along DNA to split double strands into single strands in response to ATP hydrolysis during replication. Photo credit: Shikai Jin
Knowing the structure of a complex biological system is far from enough to understand how it works. It helps to know how the system is moving.
With this in mind, researchers at Rice University have modeled a key mechanism by which DNA replicates.
By combining structural experiments and computer simulations, bioscientist Yang Gao, theoretical physicist Peter Wolynes, graduate student Shikai Jin and their colleagues have uncovered details of how helicases, a family of ring-shaped motor proteins, bring DNA into conflict during replication. Their work could reveal new targets for disease-fighting drugs.
The synergy between the experiments and large-scale simulations they describe in the Proceedings of the National Academy of Sciences could become a paradigm for modeling the mechanisms of many complex biological systems.
“These are dynamic processes that cannot be well captured by experimental methods alone,” said Gao, assistant professor of life sciences and CPRIT fellow in cancer research. “But it is important to show the mechanisms of these helicases because they are essential for DNA replication and also potential drug targets.”
Hexameric helicases have six faces that self-assemble from peptides into a disc-like ring that separates parental DNA double strands into daughter single strands. So far, the researchers have not been able to determine exactly how the helicase proceeds when unpacking the double strand.
The Rice simulations support the idea that DNA-binding loops within the six subunits of the helicase form a type of staircase that moves down the DNA backbone, powered by ATP hydrolysis, the process by which stored chemical energy in ATP molecules is released becomes.
It was known that ATP is attracted to NTPase proteins in each subunit to drive stepping motion. But researchers didn’t fully understand that ATP hydrolysis is key. The Rice team found that ATP tightly binds the helicase subunits, but that hydrolysis significantly lowers the energy barrier for subunit dissociation, allowing the protein to take a step forward.
The researchers noted that because the helicase-DNA complex is so large, there have been few attempts to simulate the translocation of the helicase from one end of the strand to the other. Rice’s hybrid of two coarse-grained simulation techniques provided an opportunity to study the process from start to finish.
The simulations uncovered several previously unknown intermediate states and revealed the interactions involved in the long-distance motion of the helicase. They showed that each translocation step can travel more than 12 nucleotides along the backbone.
To look for the mechanism, the team focused on the T7 bacteriophage, a virus that infects bacteria that are often used as a model system. To simulate its helicase, known as gp4, the researchers combined two force fields: AWSEM, originally developed by Wolynes and his colleagues to predict how proteins fold, and open3SPN2, a DNA simulator developed by the molecular engineer Juan de Pablo at the University of Chicago. Force fields describe the forces that determine how atoms and molecules move when in contact. (The novel combination of force fields was the subject of a 2021 paper led by Rice in PLOS Computational Biology.)
Both force fields are coarse-grained molecular models based on machine learning that use only a subset of the atoms in a system, but still provide accurate results while significantly reducing computation time.
“The combination of the models enabled GPU acceleration, allowing us to run our molecular dynamics simulations very quickly,” Jin said. “The combined software is now 30 times faster than versions we used for other studies.”
It helps that T7 is half the size of helicases in human cells. “In human systems there are six different polypeptide chains in the helicase, but in T7 it’s the same one that makes six copies,” he said.
“Because our new form of open3SPN2 deals with a single strand of DNA, we can analyze processes in which the normally double-stranded DNA opens, as occurs in the presence of the helicase,” said Wolynes, co-director of the Rice’s center for Theoretical Biological Physics. “The single-stranded DNA force field itself is novel, but this was just background in this project, allowing us to look at the process in detail.”
“The cryo-EM structures that we have for these essential complexes are physiologically precise, but these systems are dynamic,” Gao said. “They have to move to do their job and we still want to know a lot about how they do that.
‘This is where these computational models can go a long way, and they will certainly be adapted to other large systems to study fairly important questions,’ he said.
Researchers identify how the bacterial replicative helicase opens to start the DNA replication process
Shikai Jin et al, Computational study of the mechanism of translocation of bacteriophage T7 gp4 helicase along ssDNA, Proceedings of the National Academy of Sciences (2022). DOI: 10.1073/pnas.2202239119
Provided by Rice University
Citation: Research team models ‘washers’ that help DNA replication (2022 August 9) Retrieved August 10, 2022 from https://phys.org/news/2022-08-team-washers-dna-replicate. html
This document is protected by copyright. Except for fair trade for the purpose of private study or research, no part may be reproduced without written permission. The content is for informational purposes only.
#Research #team #models #moving #washers #DNA #replication

Leave a Comment